Antibiotic resistance has become a major challenge of our time. Common microorganisms that inhabit the skin, mouth and nares, and fecal organisms are transmitted in the hospital setting. Handwashing procedures have had limited benefit. Operating rooms are ventilated and environmentally engineered to minimize transmission intraoperatively. The patient may be immune-compromized. The organisms that are encountered have genetically adapted to the most effective antibiotics at our disposal. even with some risk of secondary toxicity in some cases.
What is Drug Resistance?
Antimicrobial resistance is the ability of microbes, such as bacteria, viruses,
parasites, or fungi, to grow in the presence of a chemical (drug) that would
normally kill it or limit its growth.
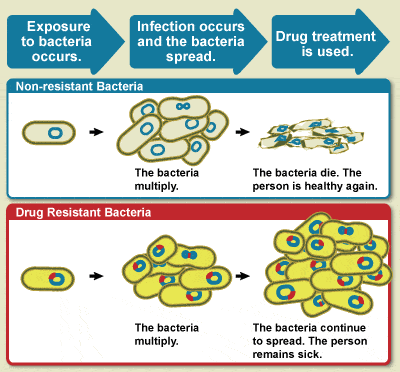
Diagram showing the difference between non-resistant bacteria and drug
resistant bacteria. Non-resistant bacteria multiply, and upon drug treatment,
the bacteria die. Drug resistant bacteria multiply as well, but upon drug
treatment, the bacteria continue to spread.
Many infectious diseases are increasingly difficult to treat because of
antimicrobial-resistant organisms, including HIV infection, staphylococcal
infection, tuberculosis, influenza, gonorrhea, candida infection, and malaria.
Between 5 and 10 percent of all hospital patients develop an infection. About
90,000 of these patients die each year as a result of their infection, up from
13,300 patient deaths in 1992.
According to the Centers for Disease Control and Prevention (April 2011),
antibiotic resistance in the United States costs an estimated $20 billion a year
in excess health care costs, $35 million in other societal costs and more than 8
million additional days that people spend in the hospital.
World Health Organization – 2014 Report
WHO/HSE/PED/AIP/2014.2
Antimicrobial resistance (AMR) is an increasingly serious threat to
global public health. AMR develops when a microorganism (bacteria,
fungus, virus or parasite) no longer responds to a drug to which it
was originally sensitive. This means that standard treatments no
longer work; infections are harder or impossible to control; the risk
of the spread of infection to others is increased; illness and hospital
stays are prolonged, with added economic and social costs; and the
risk of death is greater—in some cases, twice that of patients who
have infections caused by non-resistant bacteria. The problem is so
serious that it threatens the achievements of modern medicine. A
post-antibiotic era—in which common infections and minor
injuries can kill—is a very real possibility for the 21st century.
WHO is developing a global action plan for AMR that
will include:
• development of tools and standards for harmonized
surveillance of ABR in humans, and for integrated
surveillance in food-producing animals and the
food chain;
• elaboration of strategies for population-based
surveillance of AMR and its health and economic
impact; and
• collaboration between AMR surveillance networks
and centres to create or strengthen coordinated
regional and global surveillance.
AMR is a global health security threat that requires
action across government sectors and society as a
whole. Surveillance that generates reliable data is the
essential foundation of global strategies and public
health actions to contain AMR.
Resistance to Antibiotics: Are We in the Post-Antibiotic Era?
Alfonso J. Alanis
Archives of Medical Research 36 (2005) 697–705
Serious infections caused by bacteria that have become resistant
to commonly used antibiotics have become a major global healthcare
problem in the 21st century.
They not only are more severe and
require longer and more complex treatments, but they are also
significantly more expensive to diagnose and to treat. Antibiotic
resistance, initially a problem of the hospital setting associated
with an increased number of hospital acquired infections usually
in critically ill and immunosuppressed patients, has now extended
into the community causing severe infections difficult to diagnose
and treat. The molecular mechanisms by which bacteria have
become resistant to antibiotics are diverse and complex. Bacteria
have developed resistance to all different classes of antibiotics
discovered to date. The most frequent type of resistance is
acquired and transmitted horizontally via the conjugation
of a plasmid. In recent times new mechanisms of resistance
have resulted in the simultaneous development of resistance
to several antibiotic classes creating very dangerous multidrug
-resistant (MDR) bacterial strains, some also known as
‘‘superbugs’’. The indiscriminate and inappropriate use of
antibiotics in outpatient clinics, hospitalized patients and
in the food industry is the single largest factor leading to
antibiotic resistance. In recent years, the number of new
antibiotics licensed for human use in different parts of the
world has been lower than in the recent past. In addition,
there has been less innovation in the field of antimicrobial
discovery research and development. The pharmaceutical
industry, large academic institutions or the government are
not investing the necessary resources to produce the next
generation of newer safe and effective antimicrobial drugs.
In many cases, large pharmaceutical companies have terminated
their anti-infective research programs altogether due to economic
reasons. The potential negative consequences of all these events
are relevant because they put society at risk for the spread of
potentially serious MDR bacterial infections.
Significance: Nitric oxide (NO) produced by bacterial nitric oxide
synthase has recently been shown to protect the Gram-positive
pathogens Bacillus anthracis and Staphylococcus aureus from
antibiotics and oxidative stress. Using Bacillus subtilis as a model
system, we identified two NOS inhibitors that work in conjunction
with an antibiotic to kill B. subtilis. Moreover, comparison of inhibitor-bound crystal structures between the bacterial NOS and mammalian
NOS revealed an unprecedented mode of binding to the bacterial NOS
that can be further exploited for future structure-based drug design.
Overall, this work is an important advance in developing inhibitors
against gram-positive pathogens.
Summary: Nitric oxide (NO) produced by bacterial NOS functions as a
cytoprotective agent against oxidative stress in Staphylococcus aureus,
Bacillus anthracis, and Bacillus subtilis. The screening of several NOS-selective inhibitors uncovered two inhibitors with potential antimicrobial
properties. These two compounds impede the growth of B. subtilis under
oxidative stress, and crystal structures show that each compound exhibits
a unique binding mode. Both compounds serve as excellent leads for the
future development of antimicrobials against bacterial NOS-containing
bacteria.
Speciation of clinically significant coagulase negative Staphylococci
and their antibiotic resistant patterns in a tertiary care hospital
PR Vysakh, S Kandasamy and RM Prabhavathi
Int.J.Curr.Microbiol.App.Sci (2015) 4(1): 704-709
Human skin and mucus membrane has Coagulase Negative Staphylococci
(CoNS) as the indigenous flora. CoNS had become an important agent for
nosocomial infections accounting for about 9%. These infections are
difficult to treat because of the risk factors and the multiple drug resistance
nature of these organisms. The study was undertaken to identify the
prevalence of clinical isolates of CoNS, their speciation and to determine
the antibiotic sensitivity/resistant patterns of CoNS. A total of 490 isolates
were collected from different samples and subjected to biochemical
characterization and antimicrobial screening using conventional
microbiological methods. 165 isolates were identified as CoNS. 23% of
CoNS were isolated from blood, 30% from post-operative wound infections,
23% from pus, 18% from urine, 3% from body fluids (CSF, ascitic fluid etc)
and 3% from CVP tips. The antibiotic sensitivity revealed 81% resistance
to Penicillin,32% resistance to Cefoxitin, 27% resistance to Cefazolin,
55% resistance to Erythromycin, 22% to Clindamycin and 35% to
Cotrimoxazole and with no resistance to Vancomycin, Linezolid and
Ciprofloxacin. The increased recognition of CoNS and emergence of
drug resistance among them demonstrates the need to consider them
as a potent pathogen and to devise laboratory procedure to identify
and to determine the prevalence and antibiotic resistant patterns of CoNS.
Resistance to rifampicin: a review
Beth P Goldstein
The Journal of Antibiotics (2014) 67, 625–630
Resistance to rifampicin (RIF) is a broad subject covering not just the
mechanism of clinical resistance, nearly always due to a genetic change
in the b subunit of bacterial RNA polymerase (RNAP), but also how
studies of resistant polymerases have helped us understand the structure
of the enzyme, the intricacies of the transcription process and its role
in complex physiological pathways. This review can only scratch the
surface of these phenomena. The identification, in strains of
Escherichia coli, of the positions within b of the mutations determining
resistance is discussed in some detail, as are mutations in organisms
that are therapeutic targets of RIF, in particular Mycobacterium
tuberculosis. Interestingly, changes in the same three codons of
the consensus sequence occur repeatedly in unrelated RIF-resistant
(RIFr) clinical isolates of several different single mutation
predominates in mycobacteria. The utilization of our knowledge of
these mutations to develop rapid screening tests for detecting resistance
is briefly discussed. Cross-resistance among rifamycins has been a topic
of controversy; current thinking is that there is no difference in the
susceptibility of RNAP mutants to RIF, rifapentine and rifabutin.
Also summarized are intrinsic RIF resistance and other resistance
mechanisms.
Multi-drug resistance, inappropriate initial antibiotic therapy and
mortality in Gram negative severe sepsis and septic shock: A
retrospective cohort study
MD Zilberberg, AF Shorr, ST Micek, C Vazquez-Guillamet, MH Kollef
Critical Care 2014
Introduction The impact of in vitro resistance on initially appropriate antibiotic therapy
(IAAT) remains unclear. We elucidated the relationship between non-IAAT
and mortality, and between IAAT and multi-drug resistance (MDR) in
sepsis due to Gram-negative bacteremia (GNS).
Methods We conducted a single-center retrospective cohort study of adult intensive
care unit patients with bacteremia and severe sepsis/septic shock caused by
a gram-negative (GN) organism. We identified the following MDR pathogens:
MDR P. aeruginosa, extended spectrum beta lactamase and carbapenemase-
producing organisms. IAAT was defined as exposure within 24 hours of
infection onset to antibiotics active against identified pathogens based on
in vitro susceptibility testing. We derived logistic regression models to
examine a) predictors of hospital mortality and b) impact of MDR on
non-IAAT. Proportions are presented for categorical variables, and
median values with interquartile ranges (IQR) for continuous
variables.
Results Out of 1,064 patients with GNS, 351 (29.2%) did not survive
hospitalization. Non-survivors were older (66.5 (55, 73.5)
versus 63 (53, 72) years, P =0.036), sicker (Acute Physiology and
Chronic Health Evaluation II (19 (15, 25) versus 16 (12, 19),
P <0.001), and more likely to be on pressors (odds ratio (OR) 2.79,
95% confidence interval (CI) 2.12 to 3.68), mechanically ventilated
(OR 3.06, 95% CI 2.29 to 4.10) have MDR (10.0% versus 4.0%,
P <0.001) and receive non-IAAT (43.4% versus 14.6%, P <0.001).
In a logistic regression model, non-IAAT was an independent
predictor of hospital mortality (adjusted OR 3.87, 95% CI 2.77 to
5.41). In a separate model, MDR was strongly associated with
the receipt of non-IAAT (adjusted OR 13.05, 95% CI 7.00 to 24.31).
Conclusions
MDR, an important determinant of non-IAAT, is associated with
a three-fold increase in the risk of hospital mortality. Given the
paucity of therapies to cover GN MDRs, prevention and
development of new agents are critical.
Phenotypic and molecular characteristics of methicillin-resistant
Staphylococcus aureus isolates from Ekiti State, Nigeria
OA Olowe, OO Kukoyi, SS Taiwo, O Ojurongbe, OO Opaleye, et al.
Infection and Drug Resistance 2013:6 87–92
Introduction: The characteristics and antimicrobial resistance profiles
of Staphylococcus aureus differs according to geographical regions and
in relation to antibiotic usage. The aim of this study was to determine
the biochemical characteristics of the prevalent S. aureus from Ekiti State,
Nigeria, and to evaluate three commonly used disk diffusion methods
(cefoxitin, oxacillin, and methicillin) for the detection of methicillin
resistance in comparison with mecA gene detection by polymerase chain
reaction.
Materials and methods: A total of 208 isolates of S. aureus recovered
from clinical specimens were included in this study. Standard
microbiological procedures were employed in isolating the strains.
Susceptibility of each isolate to methicillin (5 μg), oxacillin (1 μg),
and cefoxitin (30 μg) was carried out using the modified Kirby–Bauer/
Clinical and Laboratory Standard Institute disk diffusion technique.
They were also tested against panels of antibiotics including vancomycin.
The conventional polymerase chain reaction method was used to detect
the presence of the mecA gene.
Results: Phenotypic resistance to methicillin, oxacillin, and cefoxitin
were 32.7%, 40.3%, and 46.5%, respectively. The mecA gene was detected
in 40 isolates, giving a methicillin-resistant S. aureus (MRSA) prevalence
of 19.2%. The S. aureus isolates were resistant to penicillin (82.7%) and
tetracycline (65.4%), but largely susceptible to erythromycin (78.8%
sensitive), pefloxacin (82.7%), and gentamicin (88.5%). When compared
to the mecA gene as the gold standard for MRSA detection, methicillin,
oxacillin, and cefoxitin gave sensitivity rates of 70%, 80%, and 100%,
and specificity rates of 76.2%, 69.1%, and 78.5% respectively.
Conclusion: When compared with previous studies employing mecA
polymerase chain reaction for MRSA detection, the prevalence of 19.2%
reported in Ekiti State, Nigeria in this study is an indication of gradual rise
in the prevalence of MRSA in Nigeria. A cefoxitin (30 μg) disk diffusion test
is recommended above methicillin and oxacillin for the phenotypic detection
of MRSA in clinical laboratories.
Direct sequencing for rapid detection of multidrug resistant Mycobacterium
tuberculosis strains in Morocco
F Zakham, I Chaoui, AH Echchaoui, F Chetioui, M Driss Elmessaoudi, et al.
Infection and Drug Resistance 2013:6 207–213
Background: Tuberculosis (TB) is a major public health problem with high
mortality and morbidity rates, especially in low-income countries.
Disturbingly, the emergence of multidrug resistant (MDR) and extensively
drug resistant (XDR) TB cases has worsened the situation, raising concerns
of a future epidemic of virtually untreatable TB. Indeed, the rapid diagnosis
of MDR TB is a critical issue for TB management. This study is an attempt to
establish a rapid diagnosis of MDR TB by sequencing the target fragments of
the rpoB gene which linked to resistance against rifampicin and the katG gene
and inhA promoter region, which are associated with resistance to isoniazid.
Methods: For this purpose, 133 sputum samples of TB patients from Morocco
were enrolled in this study. One hundred samples were collected from new
cases, and the remaining 33 were from previously treated patients (drug
relapse or failure, chronic cases) and did not respond to anti-TB drugs after
a sufficient duration of treatment. All samples were subjected to rpoB, katG
and pinhA mutation analysis by polymerase chain reaction and DNA sequencing.
Results: Molecular analysis showed that seven strains were isoniazid-
monoresistant and 17 were rifampicin-monoresistant. MDR TB strains were
identified in nine cases (6.8%). Among them, eight were traditionally
diagnosed as critical cases, comprising four chronic and four drug-relapse
cases. The last strain was isolated from a new case. The most recorded
mutation in the rpoB gene was the substitution TCG . TTG at codon 531
(Ser531 Leu), accounting for 46.15%. Significantly, the only mutation found
in the katG gene was at codon 315 (AGC to ACC) with a Ser315Thr amino acid
change. Only one sample harbored mutation in the inhA promoter region
and was a point mutation at the −15p position (C . T). Conclusion: The
polymerase chain reaction sequencing approach is an accurate and rapid
method for detection of drug-resistant TB in clinical specimens, and could
be of great interest in the management of TB in critical cases to adjust the
treatment regimen and limit the emergence of MDR and XDR strains.
Limiting and controlling carbapenem-resistant Klebsiella pneumoniae
L Saidel-Odes, A Borer.
Infection and Drug Resistance 2014:7 9–14
Carbapenem-resistant Klebsiella pneumoniae (CRKP) is resistant to
almost all antimicrobial agents, is associated with substantial morbidity
and mortality, and poses a serious threat to public health. The ongoing
worldwide spread of this pathogen emphasizes the need for immediate
intervention. This article reviews the global spread and risk factors for
CRKP colonization/infection, and provides an overview of the strategy
to combat CRKP dissemination.
Staphylococcus aureus – antimicrobial resistance and the immuno-
compromised child
J Chase McNeil
Infection and Drug Resistance 2014:7 117–127
Children with immunocompromising conditions represent a unique
group for the acquisition of antimicrobial resistant infections due to
their frequent encounters with the health care system, need for empiric
antimicrobials, and immune dysfunction. These infections are further
complicated in that there is a relative paucity of literature on the clinical
features and management of Staphylococcus aureus infections in
immunocompromised children. The available literature on the clinical
features, antimicrobial susceptibility, and management of S. aureus
infections in immunocompromised children is reviewed. S. aureus
infections in children with human immunodeficiency virus (HIV) are
associated with higher HIV viral loads and a greater degree of CD4 T-cell
suppression. In addition, staphylococcal infections in children with HIV
often exhibit a multidrug resistant phenotype. Children with cancer have
a high rate of S. aureus bacteremia and associated complications. Increased
tolerance to antiseptics among staphylococcal isolates from pediatric
oncology patients is an emerging area of research. The incidence of S. aureus
infections among pediatric solid organ transplant recipients varies
considerably by the organ transplanted; in general however, staphylococci
figure prominently among infections in the early post-transplant period.
Staphylococcal infections are also prominent pathogens among children
with a number of immunodeficiencies, notably chronic granulomatous
disease. Significant gaps in knowledge exist regarding the epidemiology
and management of S. aureus infection in these vulnerable children.
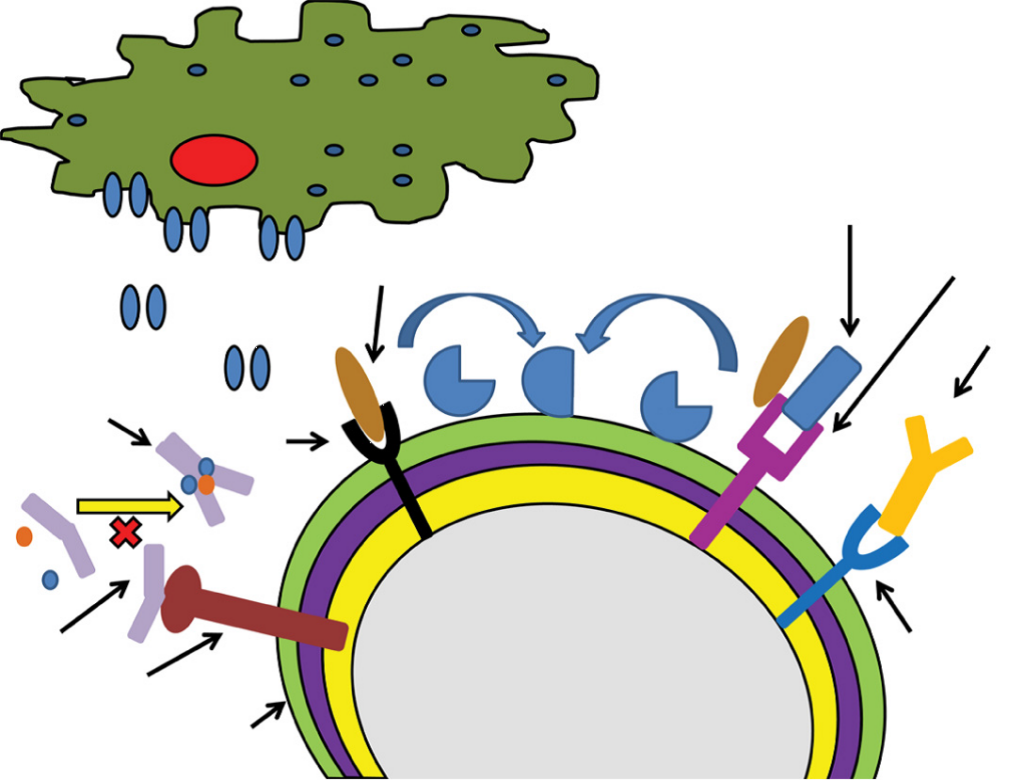
Figure 1 A schematic depiction of selected Staphylococcus aureus
mechanisms for immune evasion.
Notes: Cna interacts with C1q preventing formation of the C1qrs complex.
ClfA and SdrE each promote Factor I mediated conversion of C3b to iC3b.
Protein A is depicted binding to the Fc region of IgG preventing immunoglobulin
opsonization.
Abbreviations: ClfA, staphylococcal clumping factor A; Cna, collagen adhesin;
IgG, immunoglobulin G; PVL, Panton–Valentine leukocidin; SdrE, S. aureus
surface protein.
The Future of Antibiotics and Resistance
B Spellberg, JG Bartlett, and DN Gilbert
N Engl J Med Jan 24, 2013; 368(4): 299-302
In its recent annual report on global risks, the World Economic
Forum (WEF) concluded that
“arguably the greatest
risk . . . to human health comes in the form of antibiotic-resistant
bacteria. We live in a bacterial world where we will never be able
to stay ahead of the mutation curve. A test of our resilience is
how far behind the curve we allow ourselves to fall.”
The WEF report underscores the facts that antibiotic resistance
and the collapse of the antibiotic research and-development
pipeline continue to worsen despite our ongoing efforts on
current fronts. If we’re to develop countermeasures that
have lasting effects, new ideas that complement traditional
approaches will be needed.
Resistance is primarily the result of bacterial adaptation to eons
of antibiotic exposure. What are the fundamental implications of
this reality? First, in addition to antibiotics’ curative power, their
use naturally selects for preexisting resistant populations of bacteria
in nature. Second, it is not just “inappropriate” antibiotic use
that selects for resistance. Rather, the speed with which resistance
spreads is driven by microbial exposure to all antibiotics, whether
appropriately prescribed or not. Thus, even if all inappropriate
antibiotic use were eliminated, antibiotic-resistant infections
would still occur (albeit at lower frequency). Third, after billions
of years of evolution, microbes have most likely invented
antibiotics against every biochemical target that can be attacked
— and, of necessity, developed resistance mechanisms
to protect all those biochemical targets.
Remarkably, resistance was found even to synthetic antibiotics
that did not exist on earth until the 20th century. These results
underscore a critical reality: antibiotic resistance already exists,
widely disseminated in nature, to drugs we have not yet invented.
Interventions to Address the Antibiotic-Resistance Crisis.
Intervention Status Preventing infection
and resistance
“Self-cleaning” hospital rooms; Some commercially available
automated disinfectant application but require clinical validation;
through misting, vapor, radiation, etc. more needed
Novel drug-delivery systems to replace Basic science and
IV catheters; regenerative-tissue technology conceptual stages
to replace prosthetics; superior, noninvasive
ventilation strategies
Improvement of population health and Implementation
health care systems to reduce admissions research stage
to hospitals and skilled nursing facilities
Niche vaccines to prevent resistant Basic and clinical
bacterial infections development stage
Refilling antibiotic pipeline by aligning
economic and regulatory approaches
Models in place, expansion needed in number Government or nonprofit grants
and scope; new nonprofit corporations and contracts to defray R&D costs
needed and establish nonprofits
to develop antibiotics
Institution of novel approval pathways Proposed, legislative
(e.g., Limited Population Antibiotic and regulatory
Drug proposal) action needed
Preserving available antibiotics,
slowing resistance
Public reporting of antibiotic-use data as a Policy action needed to
basis for benchmarking and reimbursement develop and implement
Development of and reimbursement for Basic and applied research
rapid diagnostic and biomarker tests to and policy action and
enable appropriate use of antibiotics policy action needed
Elimination of use of antibiotics to Legislation proposed
promote livestock growth
New waste-treatment strategies; One strategy approaching
targeted chemical or biologic clinical trials
degradation of antibiotics in waste
Studies to define shortest effective Some trials completed
courses of antibiotics for infections
Developing microbe-attacking Preclinical, proof-of-
treatments with diminished principle stage
potential to drive resistance
Immune-based therapies, such
as infusion of monoclonal antibodies
and white cells that kill microbes
Antibiotics or biologic agents that
don’t kill bacteria but alter their ability
to trigger inflammation or cause disease
Developing treatments attacking host Preclinical, proof-of-principle stage
targets rather than microbial targets to
avoid selective pressure driving resistance
Direct moderation of host inflammation
in response to infection (e.g., cytokine
agonists or antagonists, PAMP receptor
agonists)
Sequestration of host nutrients to
prevent microbial access to nutrients
Probiotics that compete with microbial
growth
Antibiotic-Resistant Bugs Appear to Use Universal Ribosome-Stalling Mechanism
Researchers at St. Louis University say they have discovered new information
about how antibiotics like azithromycin stop staph infections, and why staph
sometimes becomes resistant to drugs. The team, led by Mee-Ngan F. Yap, Ph.D.,
believe their evidence suggests a universal, evolutionary mechanism by which
the bacteria elude this kind of drug, offering scientists a way to improve the
effectiveness of antibiotics to which bacteria have become resistant. Their
study (“Sequence selectivity of macrolide-induced translational attenuation”)
was published in PNAS.
Staphylococcus aureus is a strain of bacteria that frequently has become
resistant to antibiotics, a development that has been challenging for doctors
and dangerous for patients with severe infections. Dr. Yap and her research
team studied staph that had been treated with the antibiotic azithromycin and
learned two things: One, it turns out that the antibiotic isn’t as effective as was
previously thought. And two, the process that the bacteria use to evade the
antibiotic appears to be an evolutionary mechanism that the bacteria developed
in order to delay genetic replication when beneficial.
Genomic epidemiology of a protracted hospital outbreak caused by multidrug-
resistant Acinetobacter baumannii in Birmingham, England
MR Halachev, J Z-M Chan, CI Constantinidou, N Cumley, C Bradley, et al.
Genome Medicine 2014
Background: Multidrug-resistant Acinetobacter baumannii commonly causes
hospital outbreaks. However, within an outbreak, it can be difficult to identify
the routes of cross-infection rapidly and accurately enough to inform infection
control. Here, we describe a protracted hospital outbreak of multidrug-resistant
A. baumannii, in which whole-genome sequencing (WGS) was used to obtain
a high-resolution view of the relationships between isolates.
Methods: To delineate and investigate the outbreak, we attempted to genome-
sequence 114 isolates that had been assigned to the A. baumannii complex
by the Vitek2 system and obtained informative draft genome sequences from
102 of them. Genomes were mapped against an outbreak reference sequence
to identify single nucleotide variants (SNVs).
Results: We found that the pulsotype 27 outbreak strain was distinct from all
other genome-sequenced strains. Seventy-four isolates from 49 patients
could be assigned to the pulsotype 27 outbreak on the basis of genomic
similarity, while WGS allowed 18 isolates to be ruled out of the outbreak.
Among the pulsotype 27 outbreak isolates, we identified 31 SNVs and seven
major genotypic clusters. In two patients, we documented within-host diversity,
including mixtures of unrelated strains and within-strain clouds of SNV diversity.
By combining WGS and epidemiological data, we reconstructed potential
transmission events that linked all but 10 of the patients and confirmed links
between clinical and environmental isolates. Identification of a contaminated
bed and a burns theatre as sources of transmission led to enhanced
environmental decontamination procedures.
Conclusions: WGS is now poised to make an impact on hospital infection
prevention and control, delivering cost-effective identification of routes of
infection within a clinically relevant timeframe and allowing infection control
teams to track, and even prevent, the spread of drug-resistant hospital pathogens.
Discovery of β-lactam-resistant variants in diverse pneumococcal populations
Regine Hakenbeck
Genome Medicine 2014
Understanding of antibiotic resistance in Streptococcus pneumoniae has been
hindered by the low frequency of recombination events in bacteria and thus the
presence of large linked haplotype blocks, which preclude identification of
causative variants. A recent study combining a large number of genomes of
resistant phenotypes has given an insight into the evolving resistance to
β-lactams, providing the first large-scale identification of candidate variants
underlying resistance.
New protein detonates bacteria from within
Tel Aviv – By sequencing the DNA of bacteria resistant to viral toxins, scientists have identified novel proteins capable of stymieing growth in pathogenic, antibiotic-resistant bacteria.
Today’s arsenal of antibiotics is ineffective against some emerging strains of antibiotic-resistant pathogens. Novel inhibitors of bacterial growth therefore need to be found. One way is looking into the viruses that infect bacteria.
Key to the new initiative is the concept of fighting bacteria from within, rather than using an external chemical to batter through the bacterial cell wall. the basis of the new weapon is viral. In order to select an appropriate viral protein, researchers undertook a comprehensive screening exercise in order to identify proteins in viruses that are known to infect bacteria (bacteriophages). Bacteriophages occur abundantly in the biosphere, with different virions, genomes and lifestyles. The review was so comprehensive that it took almost three years to complete.
The screening was achieved through the use of high-throughput DNA sequencing. This is the process of determining the precise order of nucleotides within a DNA molecule. By using this advanced genetic method, the scientists identified mutations in bacterial genes that resisted the toxicity of growth inhibitors produced by bacterial viruses. Through this, a new, tiny protein was found. The protein is termed “growth inhibitor gene product (Gp) 0.6”.
Later testing found that the protein specifically targets and inhibits the activity of a protein essential to bacterial cells. The bacterial protein affected has the function of holding the microbe’s cell wall together. Without this protein functioning correctly, the cell bursts open from within and the bacterium dies.
For the next wave of research, the Israeli science group are looking further at bacterial viruses with the aim of finding compounds that facilitate improved treatment of antibiotic-resistant bacteria. Read more
Revealing bacterial targets of growth inhibitors encoded by bacteriophage T7
Shahar Molshanski-Mora, Ido Yosefa, Ruth Kiroa, Rotem Edgara, Miriam Manora, Michael Gershovitsb, Mia Lasersonb, Tal Pupkob, and Udi Qimrona,1
Author Affiliations
Edited* by Sankar Adhya, National Institutes of Health, National Cancer Institute, Bethesda, MD, and approved November 24, 2014 (received for review July 13, 2014)
Significance
Antibiotic resistance of pathogens is a growing threat to human health, requiring immediate action. Identifying new gene products of bacterial viruses and their bacterial targets may provide potent tools for fighting antibiotic-resistant strains. We show that a significant number of phage proteins are inhibitory to their bacterial host. DNA sequencing was used to map the targets of these proteins. One particular target was a key cytoskeleton protein whose function is impaired following the phage protein’s expression, resulting in bacterial death. Strikingly, in over 70 y of extensive research into the tested bacteriophage, this inhibition had never been characterized. We believe that the presented approach may be broadened to identify novel, clinically relevant bacteriophage growth inhibitors and to characterize their targets.
Abstract
Today’s arsenal of antibiotics is ineffective against some emerging strains of antibiotic-resistant pathogens. Novel inhibitors of bacterial growth therefore need to be found. The target of such bacterial-growth inhibitors must be identified, and one way to achieve this is by locating mutations that suppress their inhibitory effect. Here, we identified five growth inhibitors encoded by T7 bacteriophage. High-throughput sequencing of genomic DNA of resistant bacterial mutants evolving against three of these inhibitors revealed unique mutations in three specific genes. We found that a nonessential host gene, ppiB, is required for growth inhibition by one bacteriophage inhibitor and another nonessential gene, pcnB, is required for growth inhibition by a different inhibitor. Notably, we found a previously unidentified growth inhibitor, gene product (Gp) 0.6, that interacts with the essential cytoskeleton protein MreB and inhibits its function. We further identified mutations in two distinct regions in the mreB gene that overcome this inhibition. Bacterial two-hybrid assay and accumulation of Gp0.6 only in MreB-expressing bacteria confirmed interaction of MreB and Gp0.6. Expression of Gp0.6 resulted in lemon-shaped bacteria followed by cell lysis, as previously reported for MreB inhibitors. The described approach may be extended for the identification of new growth inhibitors and their targets across bacterial species and in higher organisms.

